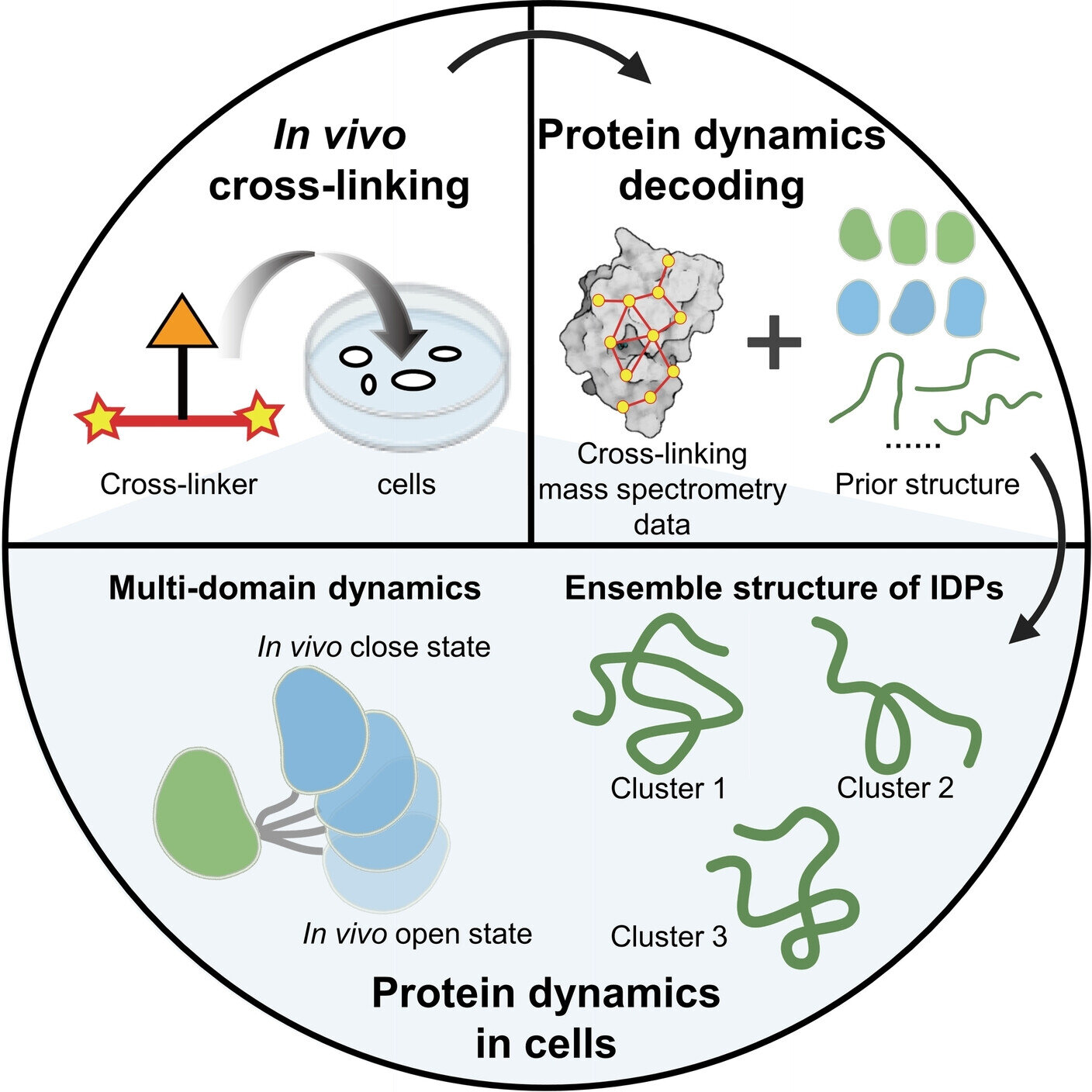
Protein dynamics are essential for a wide range of functions. The intracellular environment has a significant impact on protein dynamics, especially for intrinsically disordered proteins (IDPs).
A research group led by Prof. Zhang Lihua from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences (CAS), in collaboration with Assoc. Prof. Gong Zhou from the Precision Measurement Science and Technology Innovation Research Institute of CAS, has developed an exciting strategy to decode the dynamic structure of proteins within cells using in-vivo chemical cross-linking and mass spectrometry (in-vivo XL-MS).
In-vivo XL-MS is a promising method for analyzing protein structures within cells due to its high throughput, high sensitivity, and low requirements for protein purity.
This groundbreaking study was recently published in Angewandte Chemie International Edition on July 5.
The researchers utilized the prior structure obtained from AlphaFold2, a protein structure prediction method, as a reference. They combined in-vivo XL-MS data with various structural calculation methods to assess the compatibility between structures and cross-linking information, ultimately reconstructing the dynamic structures of various proteins within cells.
One focus of their study was on multi-domain proteins. They proposed a strategy to treat the domains as a whole, utilizing XL-MS data between the domains to model the dynamic structures of proteins within cells. They successfully characterized the dynamic structures of three multi-domain proteins: calmodulin, hnRNP A1, and hnRNP D0.
For intrinsically disordered proteins (IDPs), the researchers introduced two complementary structural characterization strategies. One strategy involved directly converting XL-MS data into distance constraints for IDP structure calculation. The other strategy employed unbiased sampling using all-atom molecular dynamics simulations, followed by evaluation and selection of the sampled structures based on XL-MS data. Using these strategies, they decoded the ensemble conformations of two highly mobile high-mobility group proteins within cells: HMG-I/Y and HMG-17.
“Our study provides technical support for a deeper understanding of the molecular mechanisms underlying protein functionality in the cellular microenvironment,” said Prof. Zhang.