Exciting Breakthrough: Low-Cost Technology for Targeted RNA Sequencing Developed by CHOP Researchers
Researchers at Children’s Hospital of Philadelphia (CHOP) have made a groundbreaking discovery that could revolutionize the field of diagnostics and treatments. They have developed a versatile and cost-effective technology called TEQUILA-seq, which enables targeted sequencing of full-length RNA molecules. This innovative technology has the potential to accelerate the discovery of new diagnostic and therapeutic solutions for a wide range of diseases.
On the journey from gene to protein, RNA molecules undergo a process called alternative splicing, where they can be cut and joined in different ways. This process allows a single gene to encode multiple proteins. However, dysregulation of alternative splicing can lead to pathogenic RNA molecules, particularly in diseases like cancer. To understand the role of alternative splicing in disease, researchers need an accurate account of all the RNA molecules that come from a single gene, known as “transcript isoforms”.
One method for achieving this is through “long-read” RNA sequencing platforms, which can sequence RNA molecules over 10,000 bases in length, capturing the entirety of the transcript isoforms. However, these platforms have limited sequencing yield, making them expensive and impractical for widespread use, especially in clinical settings. Targeted sequencing, which involves enriching specific nucleic acid sequences of interest before sequencing, is a useful strategy to enhance coverage of predefined targets. However, the cost and complexity of target capture have been major obstacles.
“TEQUILA-seq solves the problem of expensive and difficult-to-set-up technologies for targeted sequencing of full-length RNA molecules,” said co-senior author Lan Lin, Ph.D., Assistant Professor of Pathology and Laboratory Medicine at CHOP. “This technology is both inexpensive and easy to use, allowing researchers to adapt it for different purposes. They can choose which genes they want to sequence and create the reagents for target capture in their own labs. This has the potential to accelerate the discovery of new diagnostic and therapeutic solutions for various diseases.”
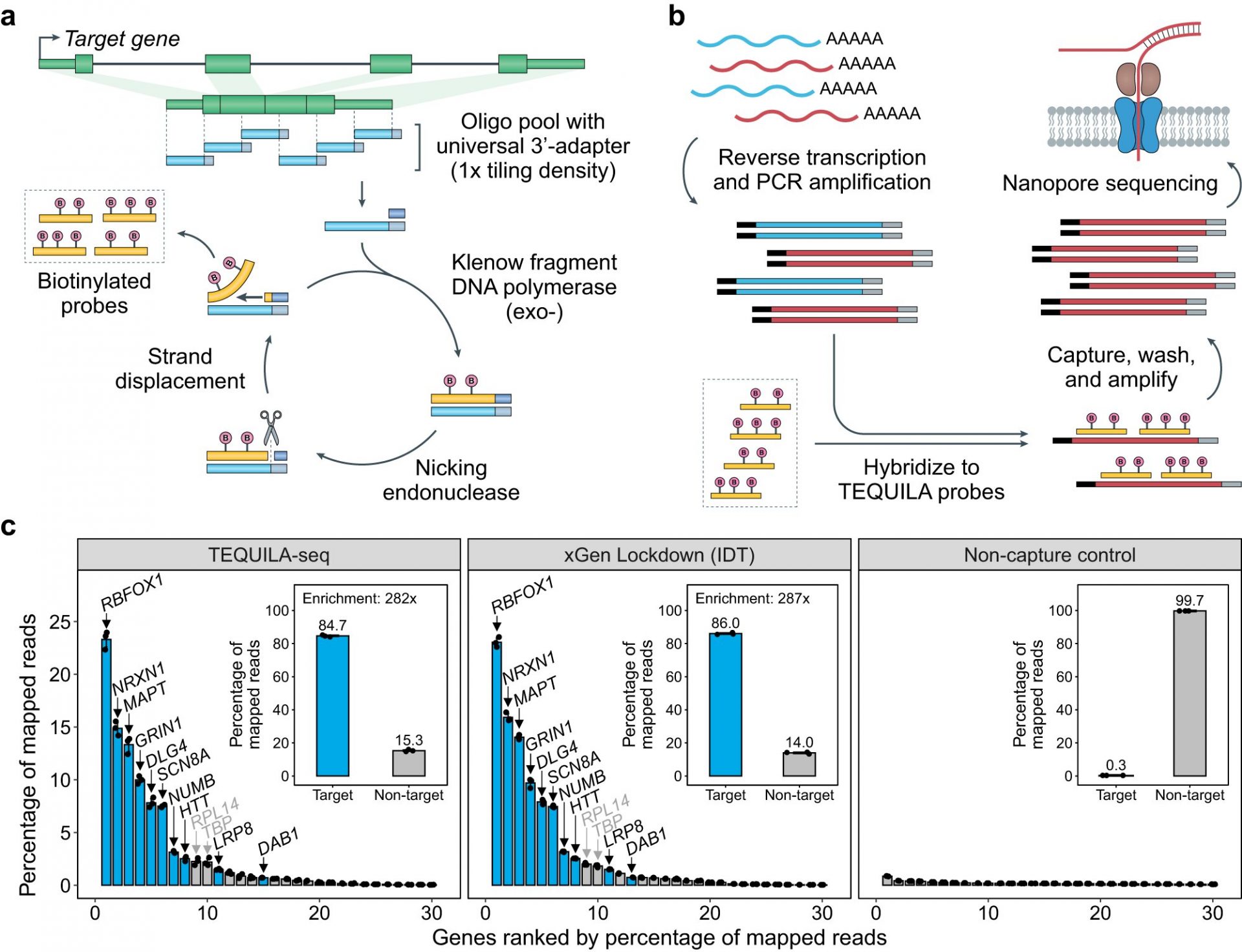
TEQUILA-seq utilizes a method called hybridization capture-based enrichment, which uses short pieces of nucleic acids called oligonucleotides as capture probes. These probes are designed to hybridize to their targets based on nucleic acid sequence complementarity, allowing for easy capture and isolation of target sequences. However, commercially synthesized biotinylated capture probes are expensive and have limited usability.
To overcome this limitation, the CHOP researchers developed TEQUILA-seq, which incorporates a novel isothermal strand displacement amplification reaction. This reaction can synthesize large quantities of biotinylated capture probes from a cheap pool of non-biotinylated oligos. With just 2 ng of template oligos, researchers can generate 25 ug of TEQUILA probes, sufficient for at least 250 capture reactions. This cost-effective strategy makes TEQUILA-seq scalable for large target panels and numerous biological samples.
In benchmarking tests, TEQUILA-seq performed as well as commercial capture probes in target capture and enrichment, while being significantly cheaper. Additionally, TEQUILA-seq enhanced detection and preserved quantification of target RNA molecules.
“TEQUILA-seq allows us to deeply sequence the full-length RNA molecules for any gene set across many biological samples,” said co-senior author Yi Xing, Ph.D., director of the Center for Computational and Genomic Medicine at CHOP. “This opens up a wide range of medical applications, from RNA-guided genetic diagnosis to therapy development.”
As a demonstration of its biomedical utility, TEQUILA-seq was used to profile full-length RNA molecules of 468 actionable cancer genes across 40 breast cancer cell lines. The researchers discovered previously unknown transcript isoforms in well-studied cancer genes, shedding light on how genes that protect against cancer are inactivated in individual tumors.
“Our work shows that TEQUILA-seq can be broadly used for targeted sequencing of full-length RNA molecules,” said Dr. Lin. “TEQUILA probes are compatible with both targeted RNA and DNA sequencing, on both long-read and short-read platforms. The ability to generate large quantities of biotinylated capture probes at a low cost and with ease can facilitate large-scale studies for many applications in basic, translational, and clinical research.”