A groundbreaking study conducted by University of Maryland physicists has unveiled new insights into the cellular processes that govern genes. Published in the esteemed journal Science Advances, the research explains how the dynamics of chromatin, a polymer that packages DNA, regulate gene expression.
Through the use of machine learning and statistical algorithms, a team of researchers led by Physics Professor Arpita Upadhyaya and National Institutes of Health Senior Investigator Gordon Hager made a groundbreaking discovery. They found that chromatin can rapidly switch between a lower and higher mobility state, with the extent of chromatin movement playing a crucial role in gene expression.
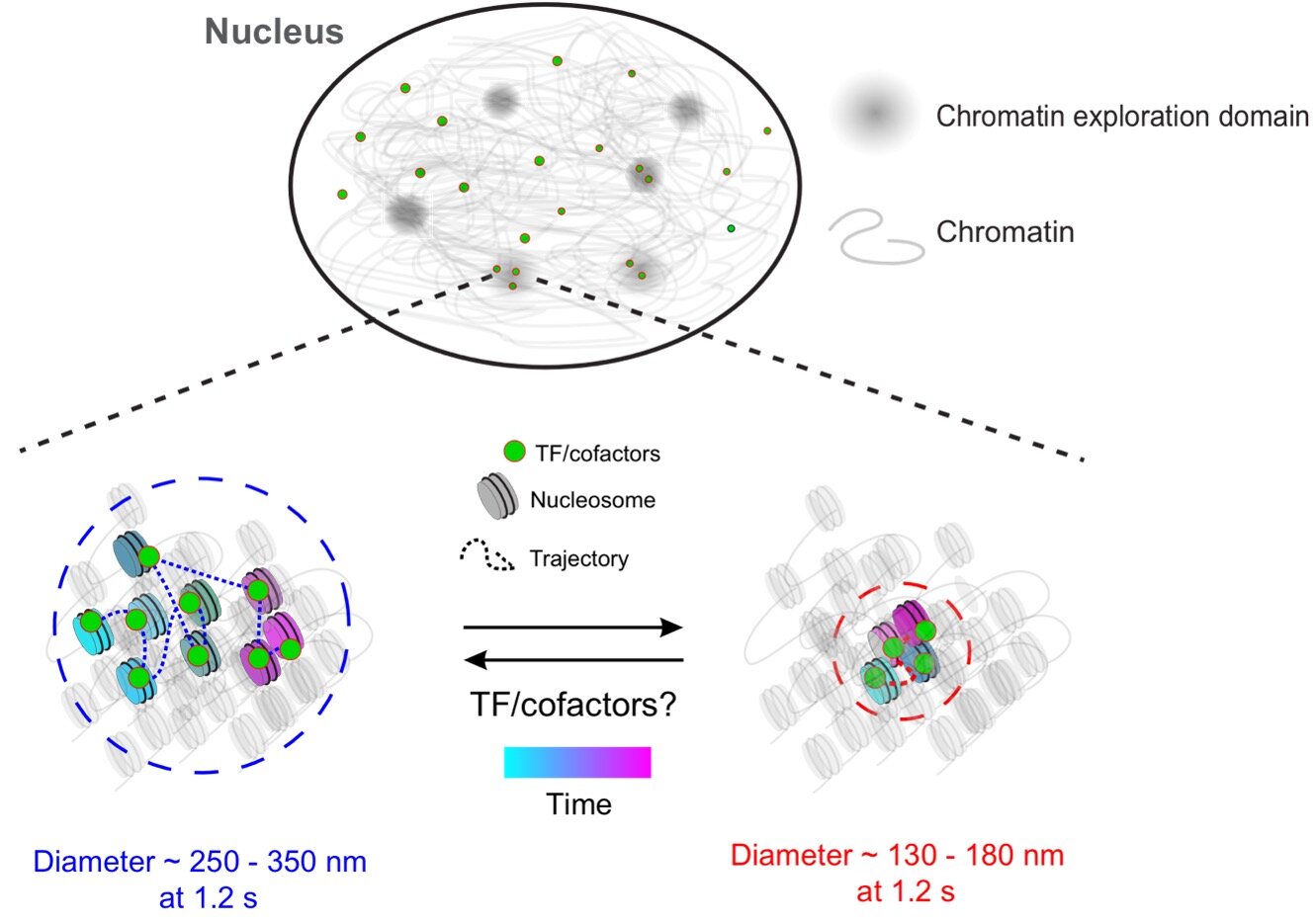
Interestingly, the study revealed that transcription factors (TFs), proteins that control gene activity, exhibit the same mobility as the chromatin they bind to. The researchers focused on a group of TFs called nuclear receptors, which are targeted by drugs used to treat various diseases and conditions.
“The nuclear receptors in our study are important therapeutic targets for breast cancer, prostate cancer, and diabetes,” explained Kaustubh Wagh (Ph.D. physics), the study’s first author. “Understanding their basic mechanism of action is essential to establish a baseline for how these proteins function.”
These findings have significant implications for medicine and could potentially lead to advancements in various fields.
On the Move
Children inherit genetic information from their parents in the form of DNA, which contains instructions for protein production. To fit inside a cell’s nucleus, DNA, which is about 2 meters long when stretched, must be compacted 100,000 times into chromatin. However, chromatin is not stationary.
“We know that the organization of the genome in our cells’ nucleus greatly impacts gene expression,” said Wagh. “However, what often goes unnoticed is that chromatin is constantly in motion within the cell, and this mobility may have significant implications for gene regulation.”
The research team, in collaboration with experts from the National Cancer Institute, the University of Buenos Aires, and the University of Southern Denmark, discovered that chromatin switches between two distinct mobility states: a lower state and a higher state. This challenges previous theories that suggested fixed chromatin mobilities in different parts of the nucleus.
“Previous studies proposed that different chromatin mobility states occupy specific regions of the cell nucleus. However, these studies were conducted on a sub-second timescale,” explained Upadhyaya, who holds a joint appointment in the Institute for Physical Science and Technology. “We expand on this model by demonstrating that the chromatin polymer can locally switch between two mobility states over longer timescales.”
The researchers also found that transcriptionally active TFs preferentially bind to chromatin in the lower mobility state. They were surprised to discover that TF molecules in this state bind for longer periods, potentially influencing gene regulation.
Discovering a Raft in the Ocean
This study significantly advances our understanding of chromatin dynamics and gene expression. The researchers plan to utilize their framework to investigate how mutations affect the function of TFs, providing insights into the development of various diseases.
“We can now determine whether a specific disease phenotype occurs due to TFs binding for excessive or insufficient periods, or not binding in the correct chromatin state,” said Wagh.
The team also aims to explore how TFs accomplish the challenging task of finding their targets. TFs target specific sequences of DNA, and only by locating and binding to these sequences can they activate nearby genes.
“A TF finding its target site is like finding a single raft in the middle of the ocean,” Upadhyaya explained. “It’s a miraculous occurrence, and we intend to uncover how it happens.”