Exciting new research from the Max Planck Institute for Dynamics and Self-Organization (MPI-DS) reveals that catalytic molecules can form metabolically active clusters through the creation and following of concentration gradients. This discovery adds a potential new mechanism to the theory of the origin of life.
The implications of this study are significant. It can help us gain a better understanding of how molecules involved in complex biological networks can form dynamic functional structures. Additionally, it provides a platform for experiments on the origins of life.
One possible scenario for the origin of life involves the spontaneous organization of interacting molecules into cell-like droplets. These molecules would then form the first self-replicating metabolic cycles, which are fundamental to all organisms. However, the traditional view suggests that the clustering of biomolecules would occur through slow and inefficient processes.
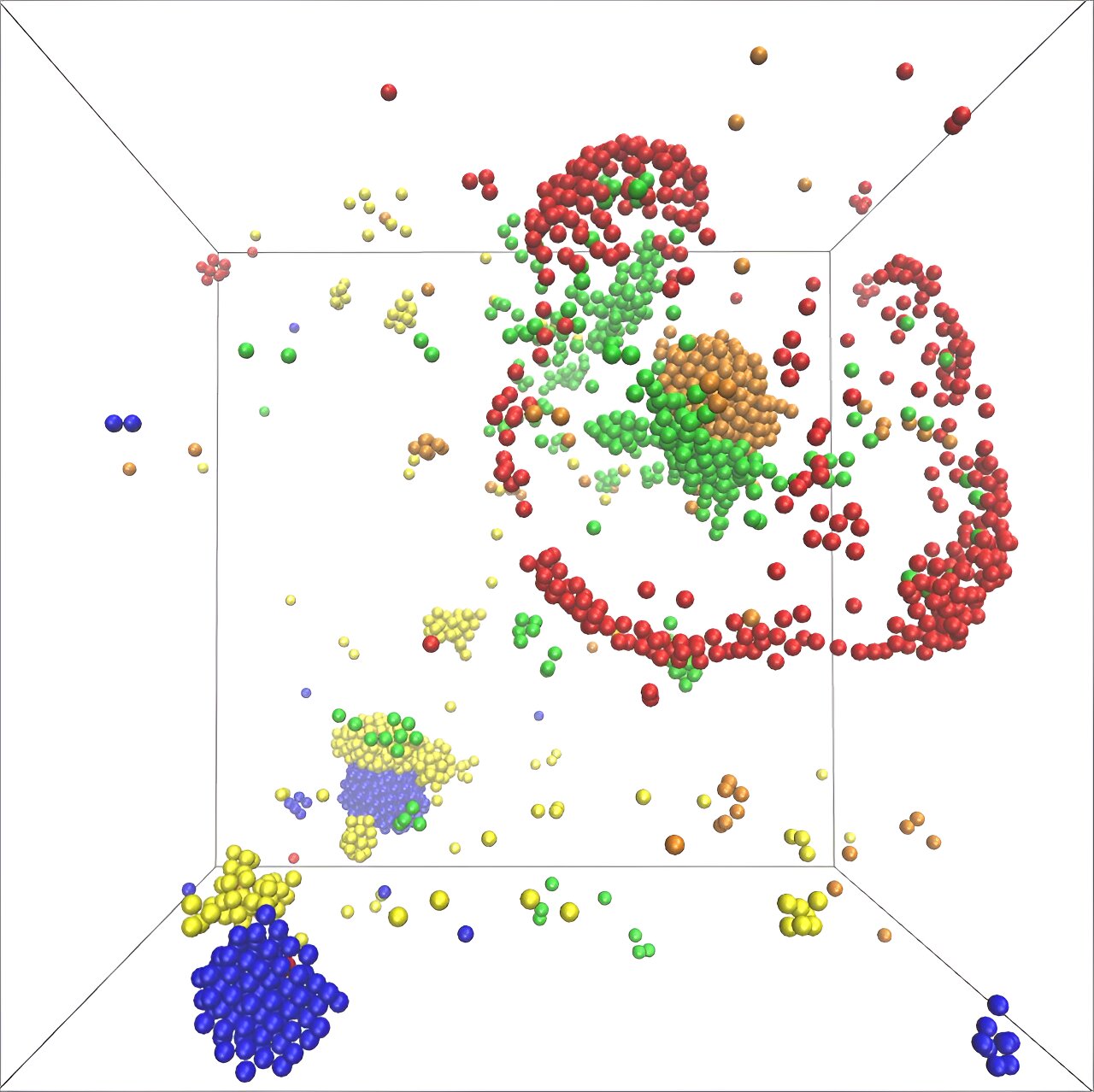
Contrary to this traditional view, the scientists at MPI-DS have proposed an alternative model that explains the fast onset of chemical reactions required for life. “For this, we considered different molecules in a simple metabolic cycle, where each species produces a chemical used by the next one,” explains Vincent Ouazan-Reboul, the first author of the study. The model takes into account the catalytic activity of the molecules, their ability to follow concentration gradients, and the order of molecules in the cycle.
The model revealed the formation of catalytic clusters consisting of various molecular species. These clusters grow exponentially fast, allowing molecules to assemble quickly and in large numbers into dynamic structures. Ramin Golestanian, director at MPI-DS, highlights the importance of the number of molecule species participating in the metabolic cycle, stating that it plays a key role in the structure of the formed clusters. The model also predicts specific advantages for odd or even numbers of participating species.
In another study, the researchers discovered that self-attraction is not necessary for clustering in a small metabolic network. Network effects can cause even self-repelling catalysts to aggregate, revealing new conditions in which complex interactions can create self-organized structures.
Overall, these groundbreaking insights add another layer to our understanding of how complex life emerged from simple molecules. They also shed light on how catalysts involved in metabolic networks can form structures. The findings have been published in the journal Nature Communications.